class: center, middle, inverse, title-slide # Multiple and logistic regression ### 2022/04/19 --- class: inverse, middle, center # Linear regression - a (brief) recap --- # Linear regression .pull-left[ ``` ## `geom_smooth()` using formula 'y ~ x' ``` 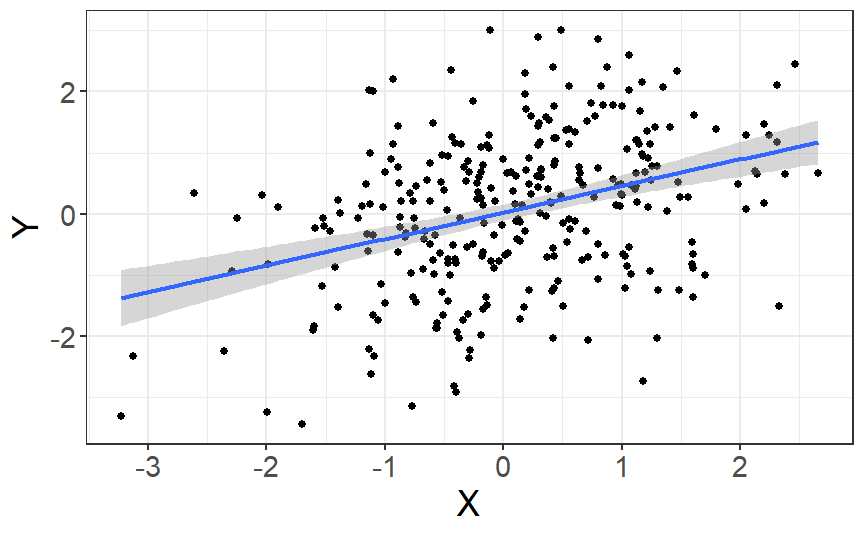 ] .pull-right[ Our job is to figure out the mathematical relationship between our *predictor(s)* and our *outcome*. $$ Y = b_0 + b_1X_i + \varepsilon_i $$ ] --- # Linear regression $$ Y = b_0 + b_1X_i + \varepsilon_i $$ --- # Linear regression $$ \color{red}Y = \color{#F6893D}{b_0} + \color{#FEBC38}{b_1}\color{blue}{X_i} + \color{green}{\varepsilon_i} $$ .red[ Y - The outcome - the dependent variable. ] .brown[ `\(b_0\)` - The *intercept*. This is the value of `\(Y\)` when `\(X\)` = 0. ] .orange[ `\(b_1\)` - The regression coefficients. This describes the steepness of the relationship between the outcome and *slope(s)*. ] .blue[ `\(X_i\)` - The predictors - our independent variables. ] .green[ `\(\varepsilon_i\)` - The *random error* - variability in our dependent variable that is not explained by our predictors. ] --- class: inverse, middle, center # Regression assumptions --- # Regression assumptions Linear regression has a number of assumptions: - Normally distributed errors - Homoscedasticity (of errors) - Independence of errors - Linearity - No perfect multicollinearity --- # Normally distributed errors .pull-left[ <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-1-1.png" width="432" /> ] .pull-right[ The *errors*, `\(\varepsilon_i\)`, are the variance left over after your model is fit. An example like that on the left is what you want to see! There is no clear pattern; the dots are evenly distributed around zero. ] --- # Skewed errors .pull-left[ <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-2-1.png" width="432" /> ] .pull-right[ In contrast, the residuals on the left are skewed. This most often happens with data that are *bounded*. For example, *reaction times* cannot be below zero; negative values are impossible. ] --- # Checking assumptions .pull-left[ The `performance` package has a very handy function called `check_model()`, which shows a variety of ways of checking the assumptions all at once. ```r library(performance) check_model(test_skew) ``` ] .pull-right[ 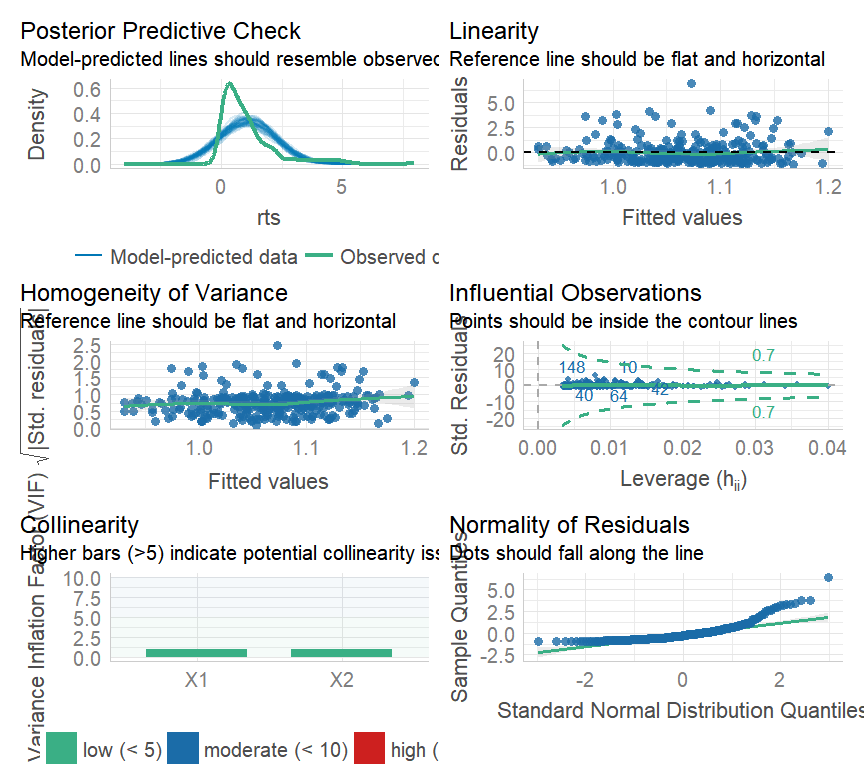 ] --- # Checking assumptions .pull-left[ You can follow up suspicious looking plots with individual functions like `check_normality()`, which uses `shapiro.test()` to check the residuals and also provides nice plots. Rely on the plots more than significance tests... ```r plot(check_normality(test_skew), type = "qq") ``` ] .pull-right[ 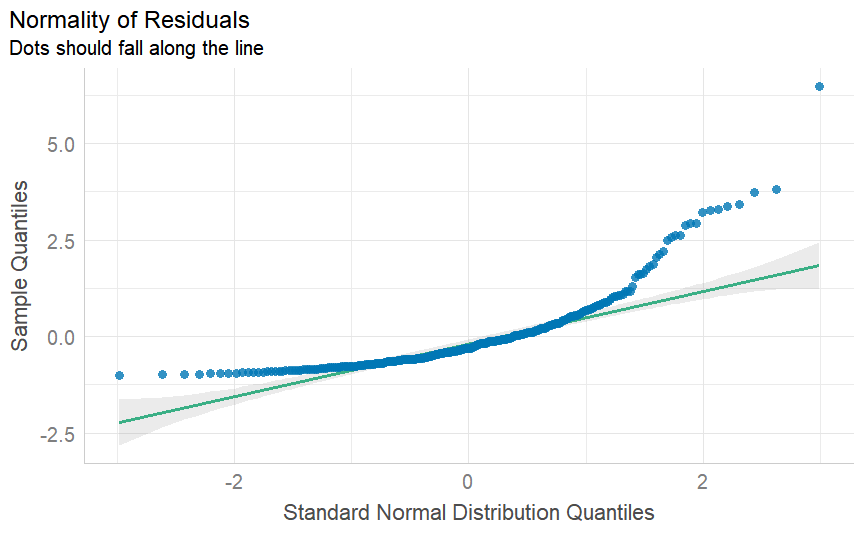 ] --- # So, about violated assumptions? 😱 .pull-left[ 1) We can think about [transformations](https://craddm.github.io/resmethods/slides/Week-23-messy-data.html?panelset3=the-data2&panelset4=with-outliers2&panelset5=right-tailed2#30) 😱 2) We could consider *non-parametric stats* - things like `wilcox.test()`, `friedman.test()`, `kruskal.test()`, all of which are based on rank transformations and thus are really more like point 1 😱 3) We should think about **why** the assumptions might be violated. Is this just part of how the data is *generated*? 🤔 ] .pull-right[ ``` ## Warning: Maximum value of original data is not included in the replicated data. ## Model may not capture the variation of the data. ``` <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-3-1.png" width="432" /> ] --- class: center, middle, inverse # Generalized Linear Models --- # Distributional families .pull-left[ <img src="3-generalized-linear-models_files/figure-html/hypothetical-norm-1.png" width="432" /> ] .pull-right[ The *normal* distribution can also be called the *Gaussian* distribution. The linear regression models we've used so far assume a *Gaussian* distribution of errors. ] --- # Generalized linear models A *Generalized Linear Model* - fit with `glm()` - allows you to specify what type of family of probability distributions the data are drawn from. .panelset[ .panel[.panel-name[The data] ```r skewed_var <- rgamma(300, 1) hist(skewed_var) ``` <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-4-1.png" width="432" /> ] .panel[.panel-name[With lm] ```r lm(skewed_var ~ X1 + X2) ``` ``` ## ## Call: ## lm(formula = skewed_var ~ X1 + X2) ## ## Coefficients: ## (Intercept) X1 X2 ## 1.01292 -0.13856 0.02078 ``` ] .panel[.panel-name[With glm] ```r glm(skewed_var ~ X1 + X2, family = "gaussian") ``` ``` ## ## Call: glm(formula = skewed_var ~ X1 + X2, family = "gaussian") ## ## Coefficients: ## (Intercept) X1 X2 ## 1.01292 -0.13856 0.02078 ## ## Degrees of Freedom: 299 Total (i.e. Null); 297 Residual ## Null Deviance: 329.7 ## Residual Deviance: 323.6 AIC: 882.1 ``` ] ] --- # Categorical outcome variables Suppose you have a *discrete*, *categorical* outcome. Examples of categorical outcomes: - correct or incorrect answer - person commits an offence or does not Examples of counts: - Number of items successfully recalled - Number of offences committed --- # The binomial distribution A coin only has two sides: heads or tails. Assuming the coin is fair, the probability - `\(P\)` - that it lands on *heads* is *.5*. So the probability it lands on *tails* - `\(1 - P\)` is also *.5*. This type of variable is called a **Bernoulli random variable**. If you toss the coin many times, the count of how many heads and how many tails occur is called a **binomial distribution**. --- # Binomial distribution If we throw the coin 100 times, how many times do we get tails? ```r coin_flips <- rbinom(n = 100, size = 1, prob = 0.5) qplot(coin_flips) ``` <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-7-1.png" width="432" /> --- # Binomial distribution What happens if we try to model individual coin flips with `lm()`? ```r coin_flips <- rbinom(n = 100, size = 1, prob = 0.5) x3 <- rnorm(100) # this is just a random variable for the purposes of demonstration! check_model(lm(coin_flips ~ x3)) ``` <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-8-1.png" width="504" /> --- # Logistic regression We get `glm()` to model a `binomial` distribution by specifying the *binomial* family. ```r coin_flips <- rbinom(n = 100, size = 1, prob = 0.5) glm(coin_flips ~ 1, family = binomial(link = "logit")) ``` ``` ## ## Call: glm(formula = coin_flips ~ 1, family = binomial(link = "logit")) ## ## Coefficients: ## (Intercept) ## -0.04001 ## ## Degrees of Freedom: 99 Total (i.e. Null); 99 Residual ## Null Deviance: 138.6 ## Residual Deviance: 138.6 AIC: 140.6 ``` --- # Logistic regression .pull-left[ **GLM with logit link** 😎 <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-11-1.png" width="432" /> ] .pull-right[ **GLM with Gaussian link** 😭 <img src="3-generalized-linear-models_files/figure-html/unnamed-chunk-12-1.png" width="432" /> ] --- # Logistic regression .pull-left[ The *logit* transformation is used to *link* our predictors to our discrete outcome variable. It helps us constrain the influence of our predictors to the range 0-1, and account for the change in *variance* with probability. ] .pull-right[ <img src="3-generalized-linear-models_files/figure-html/logit-curve-1.png" width="432" /> ] --- # Logistic regression .pull-left[ As probabilities approach zero or one, the range of possible values *decreases*. Thus, the influence of predictors on the *response scale* must also decrease as we reach one or zero. ] .pull-right[ 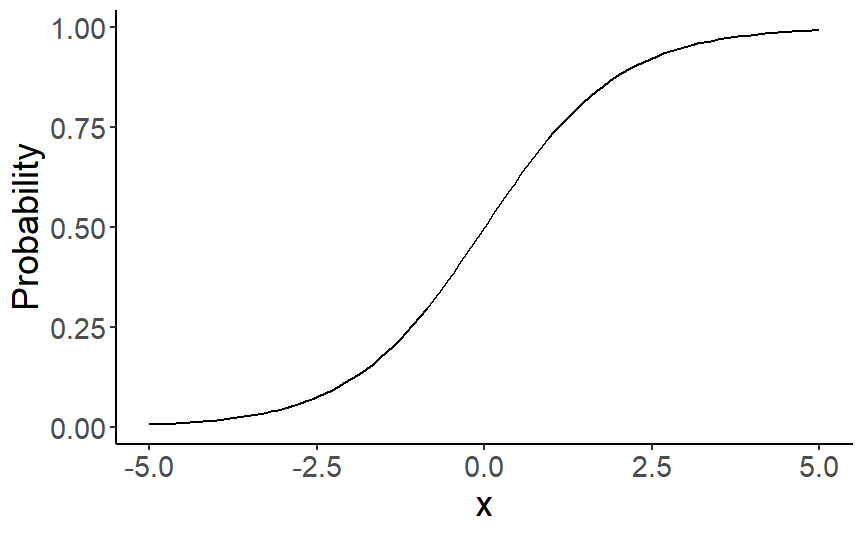 ] --- # Logistic regression $$ \color{red}{P(Y)} = \frac{1}{1 + e^-(b_0 + b_1X_1 + \varepsilon_i)} $$ .red[ `\(P(Y)\)` - The *probability* of the outcome happening. ] --- # Logistic regression $$ \color{red}{P(Y)} = \frac{\color{green}{1}}{\color{green}{1 + e^-(b_0 + b_1X_1 + \varepsilon_i)}} $$ .red[ `\(P(Y)\)` - The *probability* of the outcome happening. ] .green[ `\(\frac{1}{1 + e^-(...)}\)` - The *log-odds* (logits) of the predictors. ] --- # Odds ratios and log odds **Odds** are the ratio of one outcome versus the others. e.g. The odds of a randomly chosen day being a Friday are 1 to 6 (or 1/6 = .17) **Log odds** are the *natural log* of the odds: `$$log(\frac{p}{1-p})$$` The coefficients we get out are *log-odds* - they can be hard to interpret on their own. ```r coef(glm(coin_flips ~ 1, family = binomial(link = "logit"))) ``` ``` ## (Intercept) ## -0.04000533 ``` --- # Odds ratios and log odds If we exponeniate them, we get the *odds ratios* back. ```r exp(coef(glm(coin_flips ~ 1, family = binomial(link = "logit")))) ``` ``` ## (Intercept) ## 0.9607843 ``` So this one is roughly 1:1 heads and tails! But there's a nicer way to figure it out... --- class: inverse, middle, center # Taking penalties --  --- # Taking penalties What's the probability that a particular penalty will be scored? ``` ## PSWQ Anxious Previous Scored ## 1 18 21 56 Scored Penalty ## 2 17 32 35 Scored Penalty ## 3 16 34 35 Scored Penalty ## 4 14 40 15 Scored Penalty ## 5 5 24 47 Scored Penalty ## 6 1 15 67 Scored Penalty ``` - **PSWQ** = Penn State Worry Questionnaire - **Anxiety** = State Anxiety - **Previous** = Number of penalties scored previously --- # Taking penalties ```r pens <- glm(Scored ~ PSWQ + Anxious + Previous, family = binomial(link = "logit"), data = penalties) pens ``` ``` ## ## Call: glm(formula = Scored ~ PSWQ + Anxious + Previous, family = binomial(link = "logit"), ## data = penalties) ## ## Coefficients: ## (Intercept) PSWQ Anxious Previous ## -11.4926 -0.2514 0.2758 0.2026 ## ## Degrees of Freedom: 74 Total (i.e. Null); 71 Residual ## Null Deviance: 103.6 ## Residual Deviance: 47.42 AIC: 55.42 ``` --- ``` ## ## Call: ## glm(formula = Scored ~ PSWQ + Anxious + Previous, family = binomial(link = "logit"), ## data = penalties) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -2.31374 -0.35996 0.08334 0.53860 1.61380 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -11.49256 11.80175 -0.974 0.33016 *## PSWQ -0.25137 0.08401 -2.992 0.00277 ** ## Anxious 0.27585 0.25259 1.092 0.27480 ## Previous 0.20261 0.12932 1.567 0.11719 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 103.638 on 74 degrees of freedom ## Residual deviance: 47.416 on 71 degrees of freedom ## AIC: 55.416 ## ## Number of Fisher Scoring iterations: 6 ``` --- # The response scale and the link scale The model is fit on the *link* scale. The coefficients returned by the GLM are in *logits*, or *log-odds*. ```r coef(pens) ``` ``` ## (Intercept) PSWQ Anxious Previous ## -11.4925608 -0.2513693 0.2758489 0.2026082 ``` How do we interpret them? --- # Converting logits to odds ratios ```r coef(pens)[2:4] ``` ``` ## PSWQ Anxious Previous ## -0.2513693 0.2758489 0.2026082 ``` We can *exponentiate* the log-odds using the **exp()** function. ```r exp(coef(pens)[2:4]) ``` ``` ## PSWQ Anxious Previous ## 0.7777351 1.3176488 1.2245925 ``` --- # Odds ratios An odds ratio greater than 1 means that the odds of an outcome increase. An odds ratio less than 1 means that the odds of an outcome decrease. ```r exp(coef(pens)[2:4]) ``` ``` ## PSWQ Anxious Previous ## 0.7777351 1.3176488 1.2245925 ``` From this table, it looks like the odds of scoring a penalty decrease with increases in PSWQ but increase with increases in State Anxiety or Previous scoring rates. --- # The response scale The *response* scale is even *more* intuitive. It makes predictions using the *original* units. For a binomial distribution, that's *probabilities*. We can generate probabilities using the **predict()** function. ```r penalties$prob <- predict(pens, type = "response") head(penalties) ``` ``` ## PSWQ Anxious Previous Scored prob ## 1 18 21 56 Scored Penalty 0.7542999 ## 2 17 32 35 Scored Penalty 0.5380797 ## 3 16 34 35 Scored Penalty 0.7222563 ## 4 14 40 15 Scored Penalty 0.2811731 ## 5 5 24 47 Scored Penalty 0.9675024 ## 6 1 15 67 Scored Penalty 0.9974486 ``` --- # Model predictions Note that these model predictions *don't need to use the original data*. Let's see how the probability of scoring changes as `PSWQ` increases. .panelset[ .panel[.panel-name[Create new data] ```r new_dat <- tibble::tibble(PSWQ = seq(0, 30, by = 2), Anxious = mean(penalties$Anxious), Previous = mean(penalties$Previous)) ``` ] .panel[.panel-name[Make predictions] ```r new_dat$probs <- predict(pens, newdata = new_dat, type = "response") ``` ] .panel[.panel-name[Plot predictions] .pull-left[ ```r ggplot(new_dat, aes(x = PSWQ, y = probs)) + geom_point() + geom_line() ``` ] .pull-right[  ] ] ] --- # Model predictions Imagine you wanted to the probability of scoring for somebody with a PSWQ score of 7, an Anxious rating of 12, and a Previous scoring record of 34. .panelset[ .panel[.panel-name[Make the data] ```r new_dat <- tibble::tibble(PSWQ = 7, Anxious = 22, Previous = 34) ``` ] .panel[.panel-name[Predict log-odds] ```r predict(pens, new_dat) ``` ``` ## 1 ## -0.2947909 ``` ] .panel[.panel-name[Predict odds] ```r exp(predict(pens, new_dat)) ``` ``` ## 1 ## 0.7446873 ``` ] .panel[.panel-name[Predict probabilities] ```r predict(pens, new_dat, type = "response") ``` ``` ## 1 ## 0.4268314 ``` ] ] --- # Plotting .pull-left[ The **sjPlot** package has some excellent built in plotting tools - try the **plot_model()** function. ```r library(sjPlot) plot_model(pens, type = "pred", terms = "PSWQ") ``` ``` ## Data were 'prettified'. Consider using `terms="PSWQ [all]"` to get smooth plots. ``` ] .pull-right[  ] --- # Results tables ```r sjPlot::tab_model(pens) ``` <table style="border-collapse:collapse; border:none;"> <tr> <th style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; text-align:left; "> </th> <th colspan="3" style="border-top: double; text-align:center; font-style:normal; font-weight:bold; padding:0.2cm; ">Scored</th> </tr> <tr> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; text-align:left; ">Predictors</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">Odds Ratios</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">CI</td> <td style=" text-align:center; border-bottom:1px solid; font-style:italic; font-weight:normal; ">p</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">(Intercept)</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.00</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.00 – 64258.63</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.330</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">PSWQ</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.78</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.64 – 0.90</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; "><strong>0.003</strong></td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">Anxious</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">1.32</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.81 – 2.24</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.275</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; ">Previous</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">1.22</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.96 – 1.61</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:center; ">0.117</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm; border-top:1px solid;">Observations</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left; border-top:1px solid;" colspan="3">75</td> </tr> <tr> <td style=" padding:0.2cm; text-align:left; vertical-align:top; text-align:left; padding-top:0.1cm; padding-bottom:0.1cm;">R<sup>2</sup> Tjur</td> <td style=" padding:0.2cm; text-align:left; vertical-align:top; padding-top:0.1cm; padding-bottom:0.1cm; text-align:left;" colspan="3">0.594</td> </tr> </table> --- class: inverse, middle, center # The Titanic dataset  --- class: inverse, middle, center # The Titanic dataset  --- # The Titanic dataset ```r head(full_titanic) ``` ``` ## # A tibble: 6 x 12 ## PassengerId Survived Pclass Name Sex Age SibSp Parch Ticket Fare Cabin ## <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> ## 1 1 0 3 Braund, M~ male 22 1 0 A/5 2~ 7.25 <NA> ## 2 2 1 1 Cumings, ~ fema~ 38 1 0 PC 17~ 71.3 C85 ## 3 3 1 3 Heikkinen~ fema~ 26 0 0 STON/~ 7.92 <NA> ## 4 4 1 1 Futrelle,~ fema~ 35 1 0 113803 53.1 C123 ## 5 5 0 3 Allen, Mr~ male 35 0 0 373450 8.05 <NA> ## 6 6 0 3 Moran, Mr~ male NA 0 0 330877 8.46 <NA> ## # ... with 1 more variable: Embarked <chr> ``` Downloaded from [Kaggle](https://www.kaggle.com/c/titanic) --- # The Titanic dataset  --- # The Titanic dataset .pull-left[ ```r full_titanic %>% group_by(Survived, Sex) %>% count() ``` ``` ## # A tibble: 4 x 3 ## # Groups: Survived, Sex [4] ## Survived Sex n ## <dbl> <chr> <int> ## 1 0 female 81 ## 2 0 male 468 ## 3 1 female 233 ## 4 1 male 109 ``` ] .pull-right[ ```r full_titanic %>% group_by(Sex) %>% summarise(p = mean(Survived), Y = sum(Survived), N = n()) ``` ``` ## # A tibble: 2 x 4 ## Sex p Y N ## <chr> <dbl> <dbl> <int> ## 1 female 0.742 233 314 ## 2 male 0.189 109 577 ``` ] --- # The Titanic dataset ``` ## ## Call: ## glm(formula = Survived ~ Age + Pclass, family = binomial(), data = full_titanic) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -2.1524 -0.8466 -0.6083 1.0031 2.3929 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.296012 0.317629 7.229 4.88e-13 *** ## Age -0.041755 0.006736 -6.198 5.70e-10 *** ## Pclass2 -1.137533 0.237578 -4.788 1.68e-06 *** ## Pclass3 -2.469561 0.240182 -10.282 < 2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 964.52 on 713 degrees of freedom ## Residual deviance: 827.16 on 710 degrees of freedom ## (177 observations deleted due to missingness) ## AIC: 835.16 ## ## Number of Fisher Scoring iterations: 4 ``` --- # The Titanic dataset ```r library(emmeans) emmeans(age_class, ~Age|Pclass, type = "response") ``` ``` ## Pclass = 1: ## Age prob SE df asymp.LCL asymp.UCL ## 29.7 0.742 0.0339 Inf 0.670 0.803 ## ## Pclass = 2: ## Age prob SE df asymp.LCL asymp.UCL ## 29.7 0.480 0.0394 Inf 0.403 0.557 ## ## Pclass = 3: ## Age prob SE df asymp.LCL asymp.UCL ## 29.7 0.196 0.0216 Inf 0.157 0.241 ## ## Confidence level used: 0.95 ## Intervals are back-transformed from the logit scale ``` --- # Some final notes on Generalized Linear Models Today has focussed on **logistic** regression with *binomial* distributions. But Generalized Linear Models can be expanded to deal with many different types of outcome variable! e.g. *Counts* follow a Poisson distribution - use `family = "poisson"` Ordinal variables (e.g. Likert scale) can be modelled using *cumulative logit* models (using the **ordinal** or **brms** packages). --- #Suggested reading for categorical ordinal regression [Liddell & Kruschke (2018). Analyzing ordinal data with metric models: What could possibly go Wrong?](https://www.sciencedirect.com/science/article/pii/S0022103117307746) [Buerkner & Vuorre (2018). Ordinal Regression Models in Psychology: A Tutorial](https://psyarxiv.com/x8swp/)